Bringing a Fresh Eye to Research
 Poornima Chalasani: Modeling Molecules Poornima Chalasani: Modeling Molecules
What do accounting and computer modeling of RNA molecules have to do with one another?
Not much, says Poornima Chalasani, a master’s candidate in bioinformatics who began her college career as an accounting student. She became interested in computer engineering after a few undergraduate computing courses at Andhra University in India and completed a master’s program in computer applications in 2001.
Her initial foray into biology came after she moved to Boston University in 2002 to pursue a second master’s in computer systems engineering. “In India, biology was mostly geared toward medicine, which I was not interested in,” said Chalasani. “But in the U.S. there are many directions in biology, even for computer people.” After working as a molecular biology lab technician, she thought about how she could apply her computer expertise to the world of biological molecules.
Chalasani and a team led by Nikolay Dokholyan, her former advisor at Boston University who is now a professor at the University of North Carolina, have developed a computer simulation to compute the folding properties of RNA molecules. Their work is moving in a promising direction; in March, Chalasani won the coveted Provost’s Award at BU’s Science and Engineering Research Symposium for her part of this work.
RNA is a vital molecule, which functions as a “messenger” in the production of proteins and as a catalyst in various reactions in the cell. Like all biological molecules, RNA’s function depends on the way it’s made. “In order to understand what’s going on, we’ve got to understand the shape and composition of the tiny particles that make up our cells,” says Chalasani.
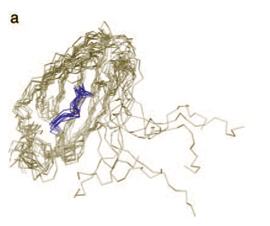
 “On the surface, the RNA molecule seems simple. But under the surface, there is a lot of complexity.” This complexity is overcome today with computers. Using powerful processors and 3-D modeling programs, the shape of RNAs can be inferred from their “sequence.” The “sequence” is a veritable alphabet, which scientists abbreviate using the first letters—A, C, U and G—of the chemicals, or nucleotides, that make up this alphabet. The programs currently used to generate 3-D models of RNA molecules are, according to Chalasani, “computationally heavy,” meaning that the algorithms in the software take a lot of computer processing power to carry out. “Some of these models take weeks to generate...our technique cuts processing time to a matter of hours,” she says. “On the surface, the RNA molecule seems simple. But under the surface, there is a lot of complexity.” This complexity is overcome today with computers. Using powerful processors and 3-D modeling programs, the shape of RNAs can be inferred from their “sequence.” The “sequence” is a veritable alphabet, which scientists abbreviate using the first letters—A, C, U and G—of the chemicals, or nucleotides, that make up this alphabet. The programs currently used to generate 3-D models of RNA molecules are, according to Chalasani, “computationally heavy,” meaning that the algorithms in the software take a lot of computer processing power to carry out. “Some of these models take weeks to generate...our technique cuts processing time to a matter of hours,” she says.
The complexity arises from the multiple interactions between atoms within the RNA itself. Each nucleotide is made up of 16 or 17 atoms, and an individual RNA strand may comprise hundreds of thousands of nucleotides.
Adopting principles used in protein computer simulations, Chalasani’s model overcomes this intrinsic complexity by consolidating sets of atoms into groups. “Instead of looking at an RNA nucleotide as a set of a few dozen atoms, we look at it as a set of three distinct groups, or ‘beads,’” Chalasani said.
The “beads” correspond to the sugar, phosphate, and base that are common structural elements in all nucleotide molecules. Looking at the atoms of RNA in terms of these groups drastically reduces the number of molecular interactions—and, thus, the computational complexity.
Once the structure is computed, the program generates a movie file. Like a digital flipbook, these movies depict how an RNA strand folds itself up into its distinct 3-dimensional shape. The group’s results have been promising.
“Of course, when you take away some of the complexity you sacrifice a bit of accuracy,” says Chalasani. “But these models do a good job of generating an accurate pathway by which an RNA molecule arrives at its unique 3-D structure.”
Chalasani says her team’s work holds promise for cell biologists and disease researchers trying to understand how mutations can lead to misshapen macromolecules that
contribute to disease.
—by Jeremy Miller |






 Poornima Chalasani: Modeling Molecules
Poornima Chalasani: Modeling Molecules  “On the surface, the RNA molecule seems simple. But under the surface, there is a lot of complexity.” This complexity is overcome today with computers. Using powerful processors and 3-D modeling programs, the shape of RNAs can be inferred from their “sequence.” The “sequence” is a veritable alphabet, which scientists abbreviate using the first letters—A, C, U and G—of the chemicals, or nucleotides, that make up this alphabet. The programs currently used to generate 3-D models of RNA molecules are, according to Chalasani, “computationally heavy,” meaning that the algorithms in the software take a lot of computer processing power to carry out. “Some of these models take weeks to generate...our technique cuts processing time to a matter of hours,” she says.
“On the surface, the RNA molecule seems simple. But under the surface, there is a lot of complexity.” This complexity is overcome today with computers. Using powerful processors and 3-D modeling programs, the shape of RNAs can be inferred from their “sequence.” The “sequence” is a veritable alphabet, which scientists abbreviate using the first letters—A, C, U and G—of the chemicals, or nucleotides, that make up this alphabet. The programs currently used to generate 3-D models of RNA molecules are, according to Chalasani, “computationally heavy,” meaning that the algorithms in the software take a lot of computer processing power to carry out. “Some of these models take weeks to generate...our technique cuts processing time to a matter of hours,” she says.