Bruce M. Boghosian
Center for Computational Science
Boston University
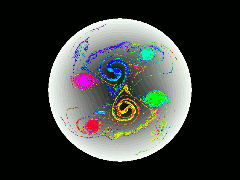
Pure-electron plasmas can be confined in cylindrically symmetric Penning traps by static electric and magnetic fields. Densities of up to 10^10 particles/cm^3, and confinement times of up to 10^4 seconds have been achieved experimentally. During the past ten years it has been appreciated that such plasmas possess equilibria that are not cylindrically symmetric. The physics governing the stability and confinement times of such e physics governing the stability and confinement times of such equilibria are not well understood.
In this work, we develop a data-parallel (CM Fortran) particle-in-cell code to model such plasmas, to gain a better understanding of their dynamical behavior. The code scatters the particles' charges to a polar coordinate grid, solves the Poisson equation for the electric field on the grid, gathers the electric field values back to the particle positions, and finally advances the particles' positions according to the guiding-center equations of motion.
Array aliasing, sorting, and segmented scan operations are used to precombine charge contributions to the same grid cell before the scatter, in order to reduce congestion in the data router, especially in the usual situation in which the particle density is strongly inhomogeneous. The Poisson solve is accomplished using the Fast Fourier Transform and the tridiagonal solver from the Connection Machine Scientific Software Library (CMSSL).
Experiment Parameters (Sequence 2)
Radial gridpoints: 256
Angular gridpoints: 256
Number of particles: 131072
Initial Mode Number: 18
Initial Mode Amplitude: 0.002
Timestep Increment: 0.01
Timesteps per Frame: 10
Video Sequences
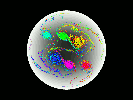
Video Sequence 1

Video Sequence 2
Still Images
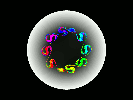


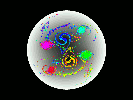

Hardware: 16-processor partition on the Thinking Machines Corporation CM-5 at Boston University.
Software: CM Fortran, C*, CMX11.
Graphics programming and video production: Erik Brisson, Scientific Computing and Visualization Group, Boston University.
Music: Richard Cornell, School for the Arts, Boston University.
Acknowledgments: Thanks to Ronsun Chu and Jonathan Wurtele for work on developing the particle-in-cell model, and to Roger Frye of the Scientific Computing and Visualization Group for programming help.
