Building a Paper Gene Circuit
New tool puts a biology lab on a piece of paper

The first case of the Ebola outbreak currently ravaging West Africa appeared in Guinea in December 2013. But it wasn’t until March 22, 2014, that scientists finally confirmed the virus as Ebola. By that point, 49 people had already died.
Why did it take so long? Partly because confirming the diagnosis required that epidemiologists fly from Europe to Africa, collect blood samples, fly back to Europe, and analyze them in sophisticated labs.
Now a team of biologists at BU, led by Professor of Biomedical Engineering and Medicine James Collins, has created a new tool that could provide a quick, cheap way to perform sophisticated lab analyses and diagnostics in the field, and may also offer a way to speed science in the lab. The tool, called a paper gene circuit, takes biological reactions out of cells and puts them onto a piece of paper. It is described in the November 6, 2014, issue of Cell.
“This could really be a game-changer for a lot of applications, including diagnostics,” says Collins, who also co-directs the Center of Synthetic Biology at BU and is a core faculty member at Harvard’s Wyss Institute. “You can literally carry this in your pocket and run an experiment in the field without any additional equipment.”
The best diagnostic tools currently use antibodies to sense things like hormones or viruses in a patient’s bloodstream. A standard pregnancy test, for example, tests for a hormone produced when a fertilized egg implants into a women’s uterus. Such tests work well but can be expensive and time-consuming to develop. “The antibody-based tests are exquisitely sensitive, and we can’t compete with that sensitivity yet,” says Keith Pardee, a postdoctoral fellow in Collins’ lab, co-author on the Cell paper, and a Wyss Institute research scientist. “But to make a custom antibody, it costs between $4,000 and $30,000, and it will take between four and six months. We made 24 different Ebola sensors and tested them in a day, for $21 each.”
The portable Ebola diagnostic is a proof-of-concept project, not yet ready for the field. But it demonstrates the power of synthetic “gene circuits” made sterile, portable, and convenient. Like computer circuits, gene circuits usually consist of a sensing component (or “input”), a logic gate, and an output, but they are crafted from parts of cells, rather than wires and transistors.
Over the past 15 years, biologists have created hundreds of these gene circuits, picking and choosing useful bits of biology and putting them together in new ways. Pardee’s circuits use a device called a “toehold switch,” created by co-author Alexander Green, also a postdoctoral fellow at BU and Pardee’s colleague at the Wyss Institute, which allows the scientists to rationally design sensors and detectors.
Because biological systems are particularly good at sensing changes in the environment—our cells constantly monitor blood sugar and scan for infection, for example—synthetic gene circuits are especially useful for detecting things like contaminants, pesticides, heavy metals, and counterfeit drugs. “You can imagine that there’s a lot of potential for these gene circuits, because they can sense and they can report by, say, changing color. Does your fruit have listeria on it? Is the soil contaminated with pesticides? The gene circuits can answer these questions,” says Pardee.
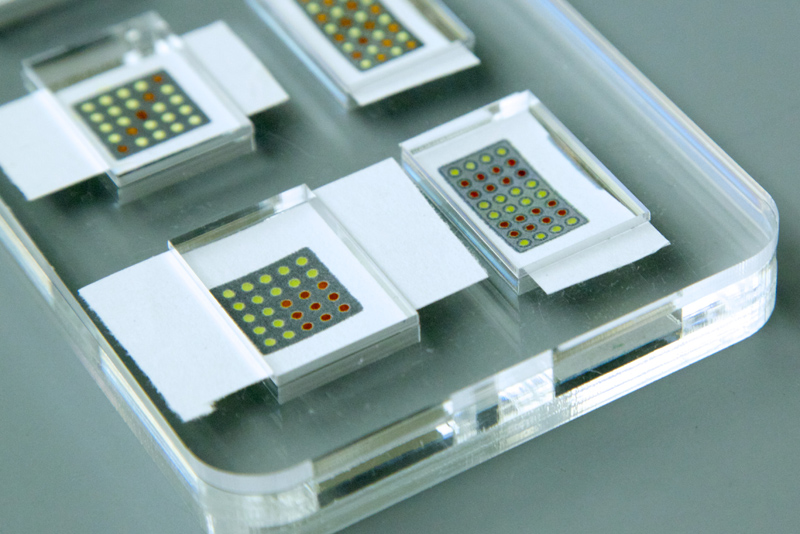
But there’s a problem. Because synthetic gene circuits are usually hosted in organisms like E. coli, they can’t be used for many applications—smearing fruit with E. coli may detect contamination, for example, but will also add to it. “For these types of applications, it’s totally inappropriate,” says Pardee. “We recognized that there was this potential but also this huge limitation.”
Pardee wondered if there was a way to build a gene circuit that could function outside a cell. Ideally, it would be sterile, easily transported, and stored without refrigeration, and it would produce answers by changing color, so a person could read the answer by eye alone. Pardee decided to try embedding the gene circuits and the necessary cellular enzymes into paper.
In concept, the idea is similar to pH strips—a chemical reaction embedded into paper that changes color when it touches an acid or a base. But would something that worked for a chemical reaction work for biology, too? “That was our first question: can we even get gene expression in paper?” says Pardee. Using a standard laser printer stocked with special wax-based inks, he printed patterns of small dots onto uncoated filter paper. Each dot served as a well or pit to hold a gene circuit and the cellular enzymes that made it work.
But Pardee wanted to take it a step further, stabilizing the components at room temperature by freeze-drying them, sticking them onto paper, then seeing if he could add water and make them work again.
“Freeze-drying is a pretty common thing to do in pharmaceuticals. If you freeze-dry a protein like insulin, you can often re-constitute it and get its function back,” says Pardee. But Pardee didn’t know if he could freeze-dry the whole package—the gene circuit instructions, the enzymes that power the reactions, the cellular machinery that builds proteins—then add water and have it pop back to its former self.
“If even one part didn’t work, then you’d be stuck. That would be the weakest link,” says Pardee. “But they all did! I was surprised. I immediately knew this was going to be really awesome.” Even better, the paper circuits still worked after a year on the shelf. “There’s still tons of work to do,” he adds. “But I knew that having that core insight was going to enable the whole thing.”
“This was Keith’s big insight—freeze-drying the circuit,” says Collins. “I couldn’t believe it worked as well as it did. We got phenomenal activity—there was basically no loss of function.”
Collins and Pardee are now working on technologies that will increase the sensitivity of the tool to make it practical for both diagnostics in the field and also for faster science in the lab. “In biology, you spend a lot of time tool building. You build your tool, then you do your experiment, then you go back to building tools,” says Pardee. “Now researchers can see if a tool works on paper in a matter of minutes and use only the best-performing tools in their experiment.”
The technology can be embedded in any porous material, such as cloth, potentially opening the door for wider applications, says Collins. He envisions smart scrubs for health care workers that can sense exposure to a virus; bandages that signal when a wound is infected with antibiotic-resistant bacteria; or smart clothing that tells a runner she’s getting dehydrated. “This opens a lot of possibilities,” says Collins. “It could give people a lot of valuable information very quickly.”

Comments & Discussion
Boston University moderates comments to facilitate an informed, substantive, civil conversation. Abusive, profane, self-promotional, misleading, incoherent or off-topic comments will be rejected. Moderators are staffed during regular business hours (EST) and can only accept comments written in English. Statistics or facts must include a citation or a link to the citation.